The wiggly targets known to scientists as “intrinsically disordered proteins” have for decades eluded capture by custom-made drugs and antibodies. But they played such important biological roles — activating opioid receptors; triggering protein misbehavior associated with neurodegeneration; killing insulin producing cells — that researchers kept after them.
Now scientists in a University of Washington lab led by Nobel laureate David Baker have cracked the challenge, using generative AI to create proteins that grab hold of the shapeshifting molecules. The discovery could unlock a suite of new drugs and diagnostic tools.
Almost half of the proteins found in humans are intrinsically disordered, “yet we’ve had no reliable way to drug [them],” Baker said in a statement. “These studies change that by giving scientists everywhere new tools for binding the unstructured half of biology.”

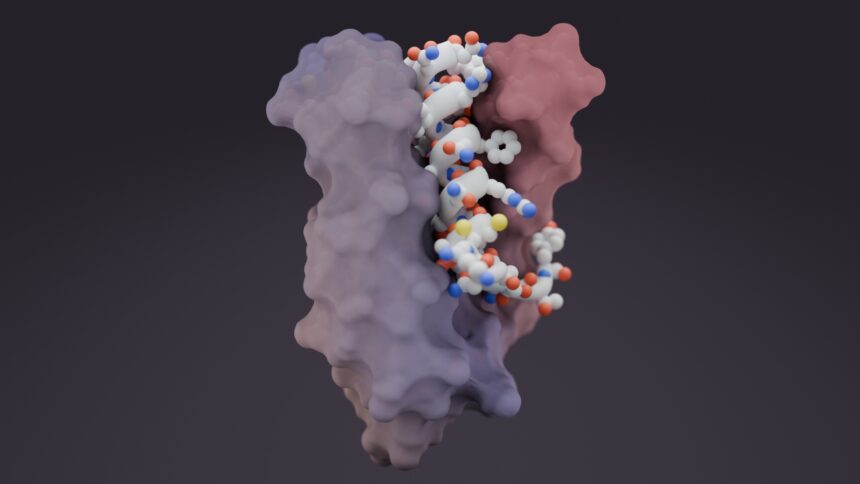
Unlike typical proteins that fold into defined, set shapes, intrinsically disordered proteins are more like cooked spaghetti — they’re floppy and lack a stable structure.
The UW scientists built a library of protein parts that can be stitched together and applied to diverse targets, zeroing in on short stretches of amino acids for binding.
The researchers tested their custom-made proteins with promising results: one successfully blocked pain signals in cultured human cells, while another dissolved protein clumps linked to type 2 diabetes. The technique also proved useful for detecting and tagging scarce but important molecules, including a disease marker screened for in newborns.
The results were striking — designer proteins successfully latched onto 39 out of 43 targets tested, a 91% success rate. Each protein folds precisely around its intended target, creating a tight, specific embrace.
The new approach to engineering proteins is described in two papers, one published today in the journal Science, and a second available as a preprint. The majority of more than two-dozen authors are from the UW’s Department of Biochemistry and the university’s Institute for Protein Design, which is led by Baker.
Researchers worldwide can access the open-source software online.

Kejia Wu, a former graduate student with the Baker lab and now a post-doctoral fellow, was a co-lead author of the newly published Science paper alongside Hanlun Jiang and Derrick Hicks. The project was exciting, Wu said, because it was so difficult and provided “space to have creative thoughts.”
Most of those ideas are nonsense and will fail, Wu said. “But then you will be able to narrow [it] down — one of your thoughts might make sense, and you just start working on it,” she continued. “So that’s the part I like the most. You’re able to come up with untraditional methods, untraditional thinking.”
And while it was difficult initially to strategize a solution for the intrinsically disordered proteins given their shapeless nature, that fluidity has an upside.
A structured, folded protein typically has just one solution for a binding protein, Wu said. “But the conformational plasticity … actually gives us freedom to target [the molecule] many different ways — and we only need one of them to work.”
Read the full article here